|
I am a final year Computer Science PhD student at University of Toronto, supervised by David Fleet and Marcus Brubaker.
I am also affiliated with Vector Institute and closely collaborate with David Lindell.
Currently, I am a research intern at Ubisoft La Forge, Toronto working on relightable and controllable head avatar, supervised by Abdallah Dib.
|

|
|
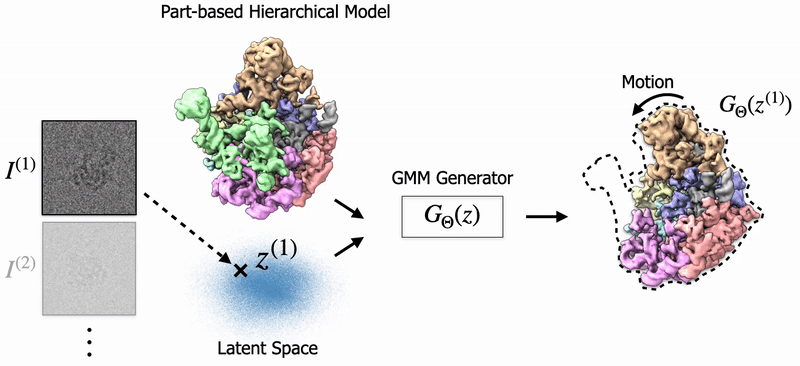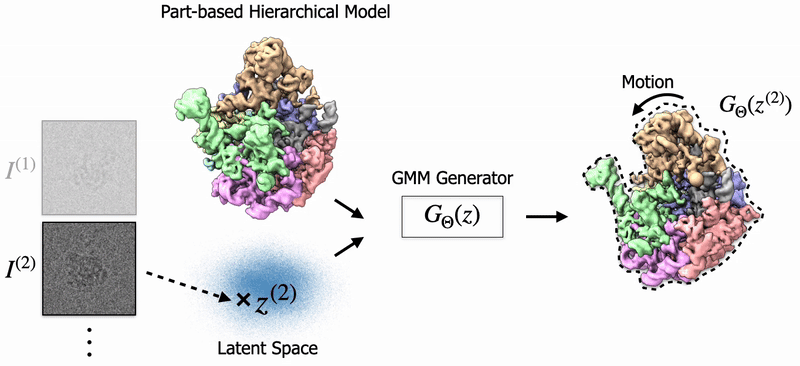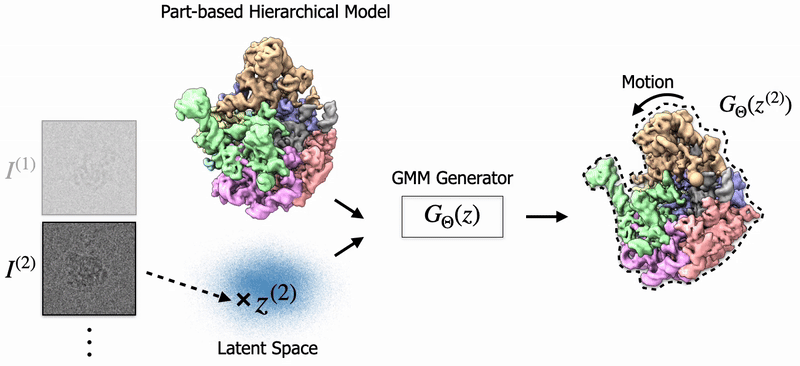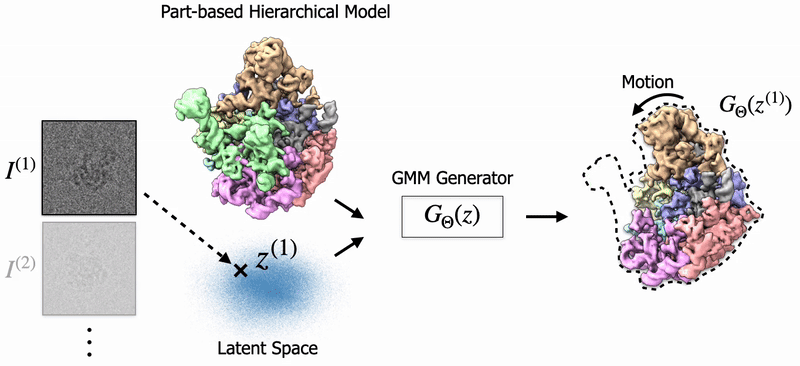
In my thesis, I studied 3D Reconstruction of biomolecules using cryo-EM, a transformational scientific imaging technique. Specifially, I've worked on Neural Fields and Gaussian Splatting to model biological structures and their dynamics. Also, I've worked on 3D Pose Estimation for cryo-EM. |
|
|
|
|
|
|
Shayan Shekarforoush, David Lindell, Marcus Brubaker, David Fleet NeurIPS 2025 arXiv / project page |
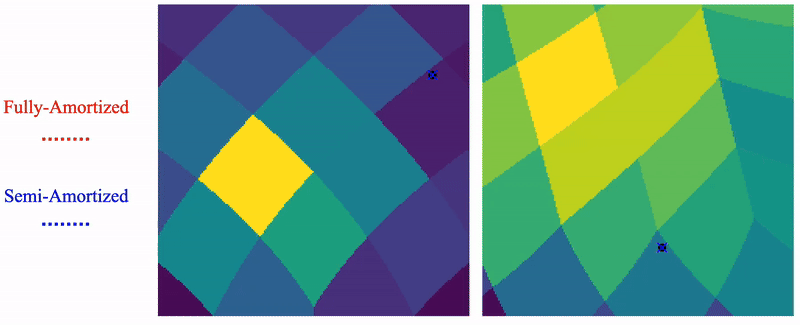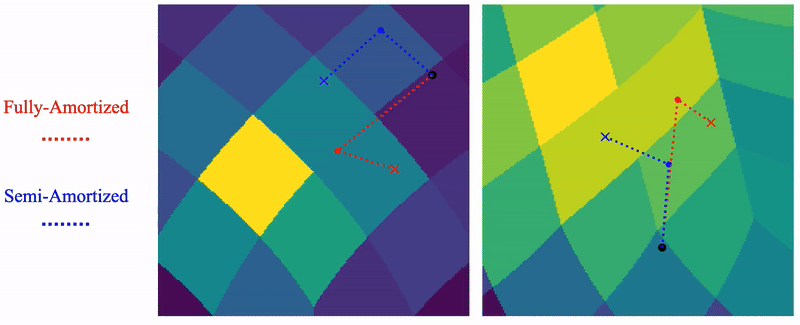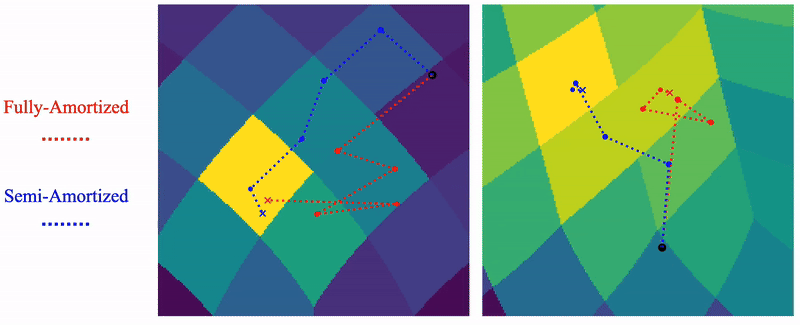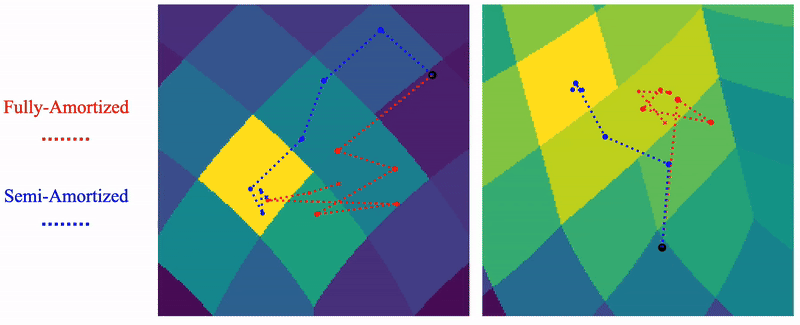
|
Shayan Shekarforoush, David Lindell, Marcus Brubaker, David Fleet NeurIPS 2024 arXiv / project page / code / Oral (MLSB Workshop) |
|
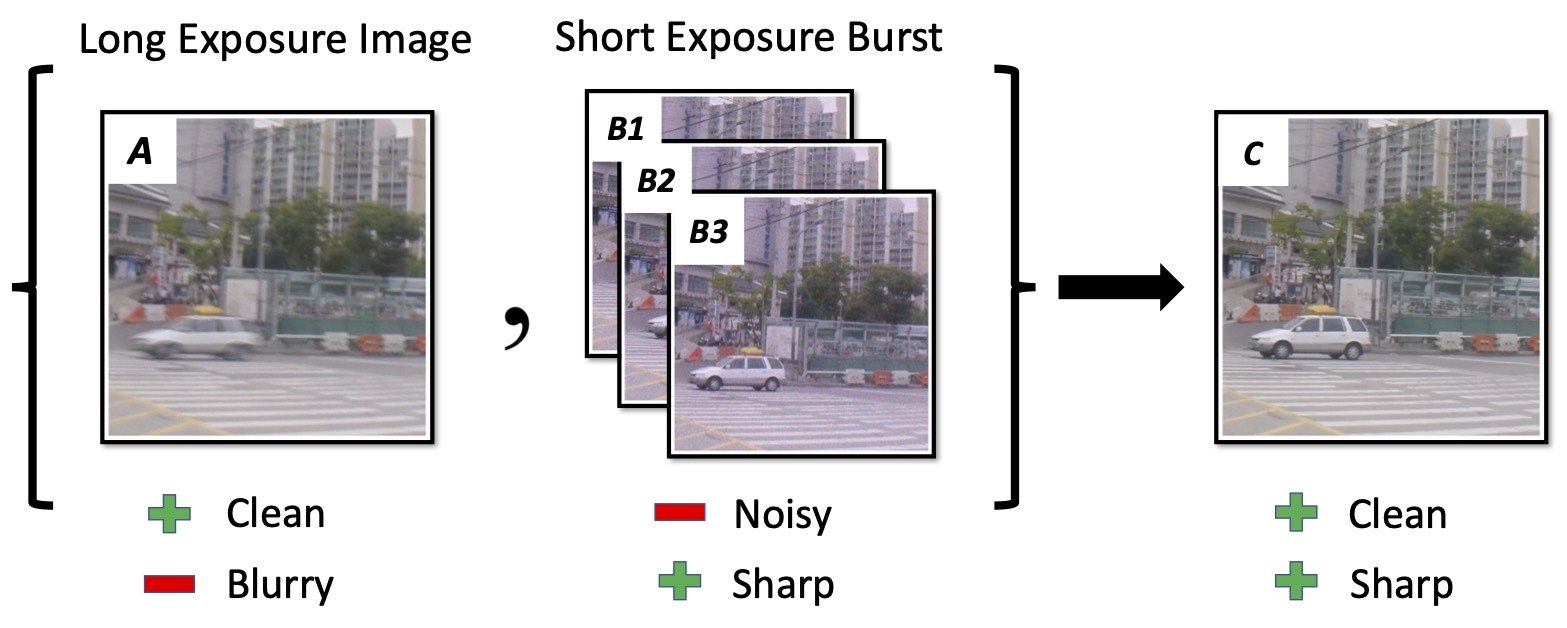
Shayan Shekarforoush, Aman Walia, Marcus Brubaker, Kosta Derpanis, Alex Levinshtein arXiv / project page |
|
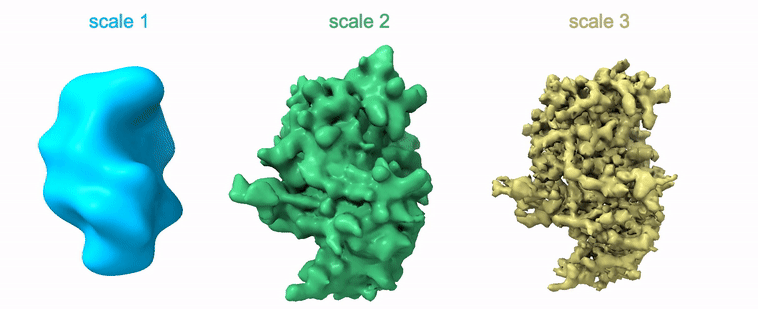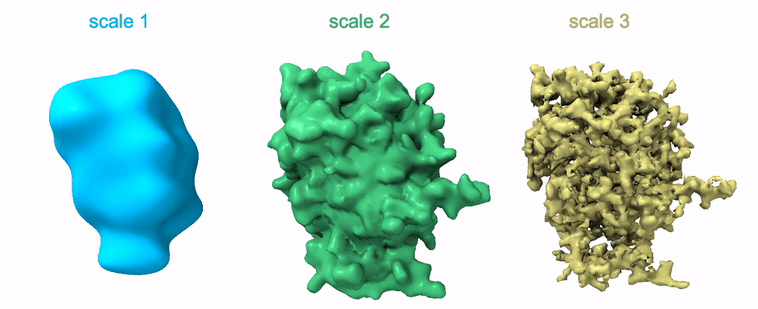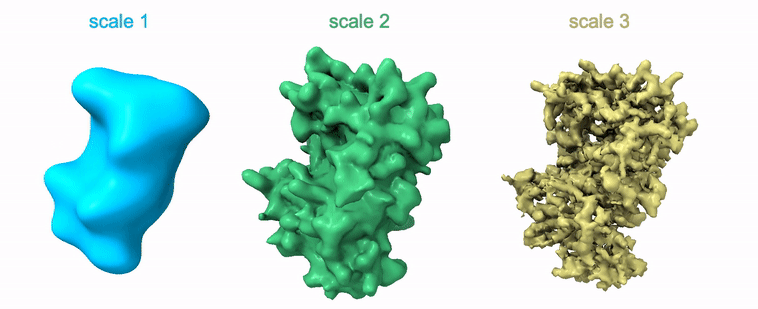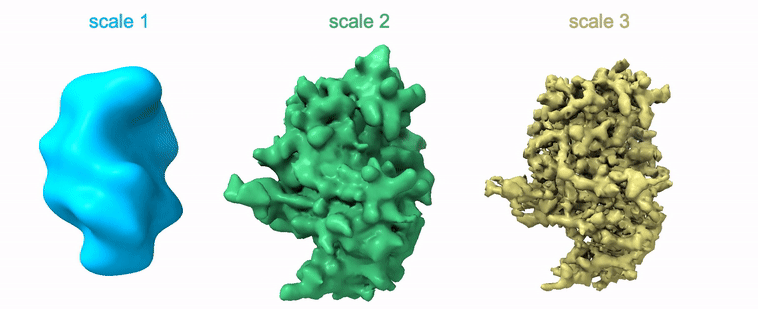
Shayan Shekarforoush, David Lindell, David Fleet, Marcus Brubaker NeurIPS 2022 arXiv / project page / code |
|
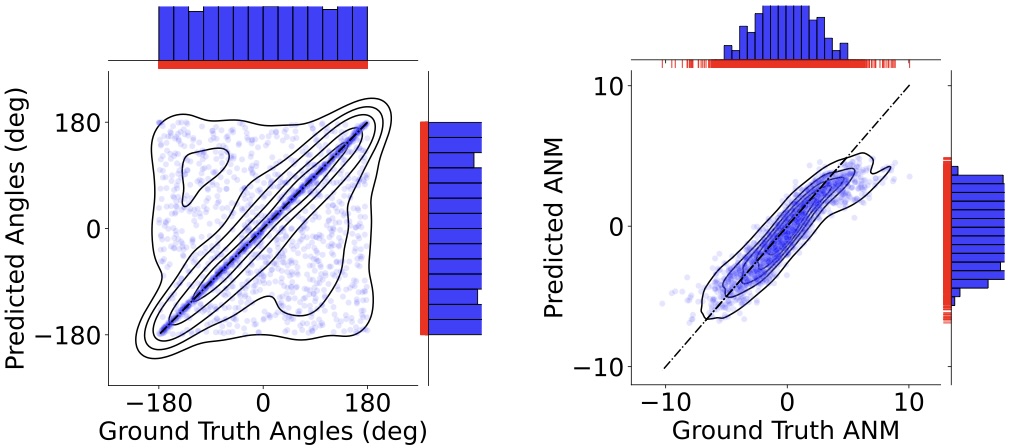
Geoffrey Woollard, Shayan Shekarforoush, Frank Wood, Marcus Brubaker, Khanh Dao Duc NeurIPS MLSB Workshop 2022 paper |
|
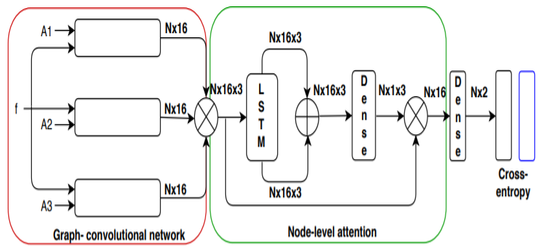
Anees Kazi, Shayan Shekarforoush, S.Arvind Krishna, Hendrik Burwinkel, Gerome Vivar, Benedict Wiestler, Karsten Kortum, Seyed-Ahmad Ahmadi, Shadi Albarqouni, Nassir Navab MICCAI 2019 paper |
|
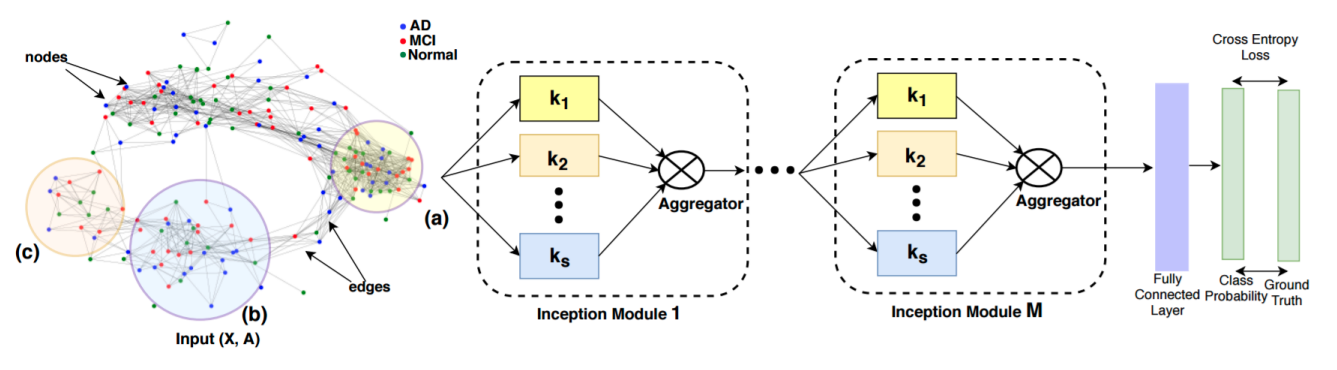
Anees Kazi, Shayan Shekarforoush, S.Arvind Krishna, Hendrik Burwinkel, Gerome Vivar, Karsten Kortuem, Seyed-Ahmad Ahmadi, Shadi Albarqouni, Nassir Navab IPMI 2019 (Oral Presentation) arXiv / code |
|
|
|
|
|
Template adapted from Jon Barron. |